Data Prediction
You are here
SLAM, IBIS and ANN – Prediction Tools
RNA-Seq has brought about a revolution in the study of gene expression. It is now possible to study the expression landscape of virtually any organism even if a species specific microarray is not readily available. However, each technological innovation brings about new problems, and for RNA-Seq, it is with the sheer quantity of data that is produced. Eyeballing it is no longer an option, and Excel will not cope. While there are powerful tools out there, they are often command-line driven, and tight deadlines mean you don't have the time to learn how to use yet another package on the command line. With Sequencher, we gave you the ability to run your NGS alignments and gene expression differential analysis through an intuitive GUI. Now with CodeLinker, we are giving you the same ability, an easy-to-use GUI with which to analyse your RNA-Seq data in depth, from your desktop.
Imagine that you are studying a disease but the expression data you have so far, while indicative, are not diagnostic. Starting with RNA-Seq differential expression results, you can use CodeLinker to find new patterns of gene expression using SLAM and ANN that are predictive of the disease you are studying. The data you have contains hidden associations (sets of genes and expression values), and with the help of SLAM/ANN, or IBIS, you can uncover these associations.
Sub-Linear Association Mining (SLAM) searches your gene expression data looking for sets of features, that is to say patterns of expression, which occur together more than might be expected by chance and discriminate between the values of any variable (gene). Once you have exposed these associations, you can then use them to train the ANN or Artificial Neural Network and then go on to classify test data. While they say that nothing good comes from a committee, that is definitely not the case when you use committees of networks. This method will generate more accurate results than would be obtained from just a single neural network. Having taught the ANN with the training data you provided, you are now ready to analyse your test data. The results can be displayed in the graphically rich Classification Plot, giving each sample in your test set a classification – predicted, true class (something different from your original classification), or unknown. This will assist in confirming or refuting your hypothesis concerning the data.
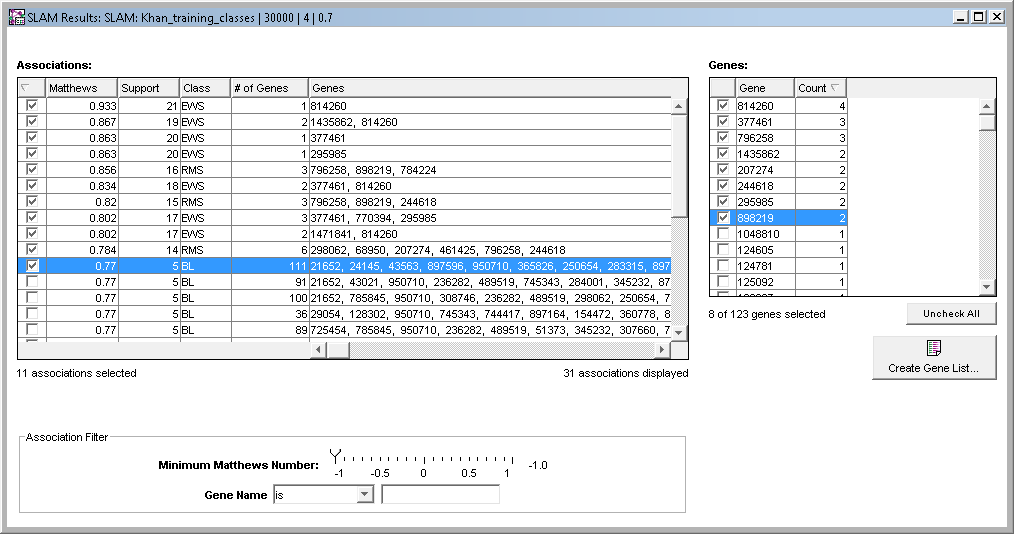
Suppose you are studying the response of certain cell types to a drug treatment. Just as you told Cuffdiff the conditions for each sample (drug dose, tissue type, phenotype), you can use the same classifications with CodeLinker. Two sets of information are imported into CodeLinker – a set of expression data, and the list of tissues and their responses to the drug treatment. These can be explored using the Integrated Bayesian Inference System (IBIS).
The IBIS classifier is a method that uses Bayesian probabilities to look at the patterns in your data. IBIS offers powerful search capabilities into your data. It can identify non-linear and combinatorial patterns of gene expression that characterize different toxicity responses, disease states, or treatment outcomes. Furthermore, it can be used to build classifiers that can identify these patterns in new samples. It can also be used as a search tool to identify single genes and small gene sets that show interesting expression patterns relative to the sample classification.
These are imported into CodeLinker, then a Linear Discriminant Analysis search is performed which evaluates the accuracy of each gene when used as linear discriminator i.e. it has the ability to separate the data into two or more classes. Genes with lower Mean Square Errors (MSEs) reflect how well the data matches the linear model. Choosing one of these, you can then display the results of the analysis on a plot whose background color gradient represents the classification that IBIS discovered; the plot displays the gene expression and spots with colors representing the initial classification that you gave. You can see with ease whether the genes you have focussed on fit the pattern you expected or are exposing new and unexpected correlations. With the 2D plot, you can explore the putative relationships between pairs of genes and find new relationships that divide along the line of your initial classification. Spotting the false positives and false negatives is as simple as looking at the colors of the spots in relation to the colored background on the graph. For more complex data where there is no linear relationship, CodeLinker provides you with the ability to perform 2D Linear Discriminant Analysis or even Quadratic or Gaussian Discriminant Analysis.